I have data.frame with two columns like the following
dat
ID Details
id_1 box1_homodomain gn=box1 os=homo sapiens p=4 se=1
id_2 sox2_plurinet gn=plu os=mus musculus p=5 se=3
I would like to split the "os=xxx" and gn="yyy" in column "Details" for all the ids and print it like following:
Id Description gn os
Îd_1 box1_homodomain box1 homo sapiens
Id_2 sox2_plurinet plu mouse musculus
I tried the using gsub approach in R but I am unable to split the os=homo sapiens and gn=box1 into their respective columns. The following R code I used
dat$gn=gsub('^[gn=][A-z][A-z]`,dat$Details)
dat$os=gsub('^[os=][A-z][A-z]`,dat$Details)
Can anyone tell me what wrong and how can it be corrected. Kindly help me.
Thanks in advance
Here's an option with tidyr:
library(tidyr)
# specify the new column names:
vars <- c("Description", "gn", "os")
# then separate the "Details" column according to regex and drop extra columns:
separate(dat, Details, into = vars, sep = "[A-Za-z]+=", extra = "drop")
# ID Description gn os
#1 id_1 box1_homodomain box1 homo sapiens
#2 id_2 sox2_plurinet plu mus musculus
1) sub and gsub To do it using sub and gsub as in the question try this. Note that each regular expressions should match all of dat$Details so that when we replace it with the capture group only the capture group remains. For the dat$GO as in the comments to the question, we remove everything up to but not including P:, replace all occurrences of ;P with a comma and remove P: and also remove semicolon and everything thereafter. Similarly for F and C:
data.frame(dat[1],
Description = sub(" .*", "", dat$Details),
gn = sub(".*gn=(.*) os=.*", "\\1", dat$Details),
os = sub(".*os=(.*) p=.*", "\\1", dat$Details),
P = gsub("P:|;.*", "", gsub(";P:", ",", sub(".*?P:", "P:", dat$GO))),
F = gsub("F:|;.*", "", gsub(";F:", ",", sub(".*?F:", "F:", dat$GO))),
C = gsub("C:|;.*", "", gsub(";C:", ",", sub(".*?C:", "C:", dat$GO))))
giving:
ID Description gn os P F C
1 id_1 box1_homodomain box1 homo sapiens p_1,p_2 F_1 C_1,C_2
2 id_2 sox2_plurinet plu mus musculus p_1 F_1,F_2 C_1
2) read.pattern Processing of dat$Details is a bit easier with read.pattern (link) in the gsubfn package as one can define a single regular expression whose capture groups represent the fields of interest. Processing of dat$GO can be simplified too by extracting the P:... fields using strapplyc (link) and then concatenating them together with paste (and similarly with the F and C fields):
library(gsubfn)
Sub <- function(string, pat) sapply(strapplyc(string, pat), paste, collapse = ",")
DF <- read.pattern(text = as.character(dat$Details),
pattern = "(.*) gn=(.*) os=(.*) p=",
col.names = c("Description", "gn", "os"),
as.is = TRUE)
cbind(dat[1], DF,
P = Sub(dat$GO, "P:(.*?);"),
F = Sub(dat$GO, "F:(.*?);"),
C = Sub(dat$GO, "C:(.*?);"))
giving:
ID Description gn os P F C
1 id_1 box1_homodomain box1 homo sapiens p_1,p_2 F_1 C_1,C_2
2 id_2 sox2_plurinet plu mus musculus p_1 F_1,F_2 C_1
Here is a visualization of the regular expression used in read.pattern:
(.*) gn=(.*) os=(.*) p=
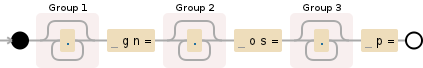
Debuggex Demo
Notes
1) If the dat$Details column is already character we could omit as.character. We could also omit as.is=TRUE if its ok to have factor columns in the result.
2) the sample output in the question has mouse but the input has mus. We have assumed it should be mus in both cases.
3) We used this for dat:
dat <-
structure(list(ID = c("id_1", "id_2"),
Details = c("box1_homodomain gn=box1 os=homo sapiens p=4 se=1",
"sox2_plurinet gn=plu os=mus musculus p=5 se=3"),
GO = c("P:p_1;P:p_2;F:F_1;C:C_1;C:C_2; ",
"P:p_1;F:F_1;F:F_2;C:C_1;")), .Names = c("ID", "Details",
"GO"), class = "data.frame", row.names = c(NA, -2L))
In the future please post the result of dput(dat) in the question.
If you love us? You can donate to us via Paypal or buy me a coffee so we can maintain and grow! Thank you!
Donate Us With