What's the easiest way to read text from a printed data.frame into a data.frame when there are string values containing spaces that interfere with read.table? For instance, this data.frame excerpt does not pose a problem:
candname party elecVotes
1 BarackObama D 365
2 JohnMcCain R 173
I can paste it into a read.table call without a problem:
dat <- read.table(text = " candname party elecVotes
1 BarackObama D 365
2 JohnMcCain R 173", header = TRUE)
But if the data has strings with spaces like this:
candname party elecVotes
1 Barack Obama D 365
2 John McCain R 173
Then read.table throws an error as it interprets "Barack" and "Obama" as two separate variables.
Read the file into L, remove the row numbers and use sub with the indicated regular expression to insert commas between the remaining fields. (Note that "\\d" matches any digit and "\\S" matches any non-whitespace character.) Now re-read it using read.csv:
Lines <- " candname party elecVotes
1 Barack Obama D 365
2 John McCain R 173"
# L <- readLines("myfile") # read file; for demonstration use next line instead
L <- readLines(textConnection(Lines))
L2 <- sub("^ *\\d+ *", "", L) # remove row numbers
read.csv(text = sub("^ *(.*\\S) +(\\S+) +(\\S+)$", "\\1,\\2,\\3", L2), as.is = TRUE)
giving:
candname party elecVotes
1 Barack Obama D 365
2 John McCain R 173
Here is a visualization of the regular expression:
^ *(.*\S) +(\S+) +(\S+)$
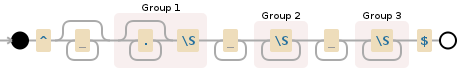
Debuggex Demo
If you love us? You can donate to us via Paypal or buy me a coffee so we can maintain and grow! Thank you!
Donate Us With