I'm working on a data science project for my intro to Data Science class, and we've decided to tackle a problem relating to desalination plants in california: "Where should we place k plants to minimize the distance to zip codes?"
The data that we have so far is, zip, city, county, pop, lat, long, amount of water.
The issue is, I can't find any resources on how to force the centroid to be constrained to staying on the coast. What we've thought of so far is: Use a normal kmeans algorithm, but move the centroid to the coast once clusters have settled (bad) Use a normal kmeans algorithm with weights, making the coastal zips have infinite weight (I've been told this isn't a great solution)
What do you guys think?
K-means is a centroid-based algorithm, or a distance-based algorithm, where we calculate the distances to assign a point to a cluster. In K-Means, each cluster is associated with a centroid.
A centroid is the imaginary or real location representing the center of the cluster. Every data point is allocated to each of the clusters through reducing the in-cluster sum of squares.
Does centroid initialization affect K means Algorithm? Yes, the final results of the k means algorithm depend on the centroid initialization as poor initialization can cause the algorithm to get stuck into an inferior local minimum.
Updating Centroids Because the initial centroids were chosen arbitrarily, your model the updates them with new cluster values. The new value might or might not occur in the dataset, in fact, it would be a coincidence if it does.
K-means does not minimize distances.
It minimizes squared errors, which is quite different. The difference is roughly that of the median, and the mean in 1 dimensional data. The error can be massive.
Here is a counter example, assuming we have the coordinates:
-1 0
+1 0
0 -1
0 101
The center chosen by k-means would be 0,25. The optimal location is 0,0. The sum of distances by k-means is > 152, the optimum location has distance 104. So here, the centroid is almost 50% worse than the optimum! But the centroid (= multivariate mean) is what k-means uses!
This is one variant how "k-means is sensitive to outliers".
It does not get better if you try to constrain it to place "centers" on the coast only...
Also, you may want to at least use Haversine distance, because in California, 1 degree north != 1 degree east, because it's not at the Equator.
Furthermore, you likely should not make the assumption that every location requires its own pipe, but rather they will be connected like a tree. This greatly reduces the cost.
I strongly suggest to treat this as a generic optimization problem, rather than k-means. K-means is an optimization too, but it may optimize the wrong function for your problem...
I would approach this by setting possible points that could be centers, i.e. your coastline.
I think this is close to Nathaniel Saul's first comment.
This way, for each iteration, instead of choosing a mean, a point out of the possible set would be chosen by proximity to the cluster.
I’ve simplified the conditions to only 2 data columns (lon. and lat.) but you should be able to extrapolate the concept. For simplicity, to demonstrate, I based this on code from here.
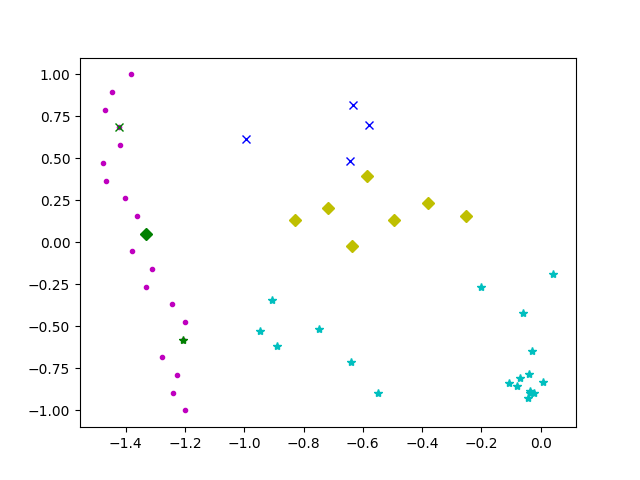
#! /usr/bin/python3.6
# Code based on:
# https://datasciencelab.wordpress.com/2013/12/12/clustering-with-k-means-in-python/
import matplotlib.pyplot as plt
import numpy as np
import random
##### Simulation START #####
# Generate possible points.
def possible_points(n=20):
y=list(np.linspace( -1, 1, n ))
x=[-1.2]
X=[]
for i in list(range(1,n)):
x.append(x[i-1]+random.uniform(-2/n,2/n) )
for a,b in zip(x,y):
X.append(np.array([a,b]))
X = np.array(X)
return X
# Generate sample
def init_board_gauss(N, k):
n = float(N)/k
X = []
for i in range(k):
c = (random.uniform(-1, 1), random.uniform(-1, 1))
s = random.uniform(0.05,0.5)
x = []
while len(x) < n:
a, b = np.array([np.random.normal(c[0], s), np.random.normal(c[1], s)])
# Continue drawing points from the distribution in the range [-1,1]
if abs(a) < 1 and abs(b) < 1:
x.append([a,b])
X.extend(x)
X = np.array(X)[:N]
return X
##### Simulation END #####
# Identify points for each center.
def cluster_points(X, mu):
clusters = {}
for x in X:
bestmukey = min([(i[0], np.linalg.norm(x-mu[i[0]])) \
for i in enumerate(mu)], key=lambda t:t[1])[0]
try:
clusters[bestmukey].append(x)
except KeyError:
clusters[bestmukey] = [x]
return clusters
# Get closest possible point for each cluster.
def closest_point(cluster,possiblePoints):
closestPoints=[]
# Check average distance for each point.
for possible in possiblePoints:
distances=[]
for point in cluster:
distances.append(np.linalg.norm(possible-point))
closestPoints.append(np.sum(distances)) # minimize total distance
# closestPoints.append(np.mean(distances))
return possiblePoints[closestPoints.index(min(closestPoints))]
# Calculate new centers.
# Here the 'coast constraint' goes.
def reevaluate_centers(clusters,possiblePoints):
newmu = []
keys = sorted(clusters.keys())
for k in keys:
newmu.append(closest_point(clusters[k],possiblePoints))
return newmu
# Check whether centers converged.
def has_converged(mu, oldmu):
return (set([tuple(a) for a in mu]) == set([tuple(a) for a in oldmu]))
# Meta function that runs the steps of the process in sequence.
def find_centers(X, K, possiblePoints):
# Initialize to K random centers
oldmu = random.sample(list(possiblePoints), K)
mu = random.sample(list(possiblePoints), K)
while not has_converged(mu, oldmu):
oldmu = mu
# Assign all points in X to clusters
clusters = cluster_points(X, mu)
# Re-evaluate centers
mu = reevaluate_centers(clusters,possiblePoints)
return(mu, clusters)
K=3
X = init_board_gauss(30,K)
possiblePoints=possible_points()
results=find_centers(X,K,possiblePoints)
# Show results
# Show constraints and clusters
# List point types
pointtypes1=["gx","gD","g*"]
plt.plot(
np.matrix(possiblePoints).transpose()[0],np.matrix(possiblePoints).transpose()[1],'m.'
)
for i in list(range(0,len(results[0]))) :
plt.plot(
np.matrix(results[0][i]).transpose()[0], np.matrix(results[0][i]).transpose()[1],pointtypes1[i]
)
pointtypes=["bx","yD","c*"]
# Show all cluster points
for i in list(range(0,len(results[1]))) :
plt.plot(
np.matrix(results[1][i]).transpose()[0],np.matrix(results[1][i]).transpose()[1],pointtypes[i]
)
plt.show()
Edited to minimize total distance.
If you love us? You can donate to us via Paypal or buy me a coffee so we can maintain and grow! Thank you!
Donate Us With