I need to colour datapoints that are outside of the the confidence bands on the plot below differently from those within the bands. Should I add a separate column to my dataset to record whether the data points are within the confidence bands? Can you provide an example please?
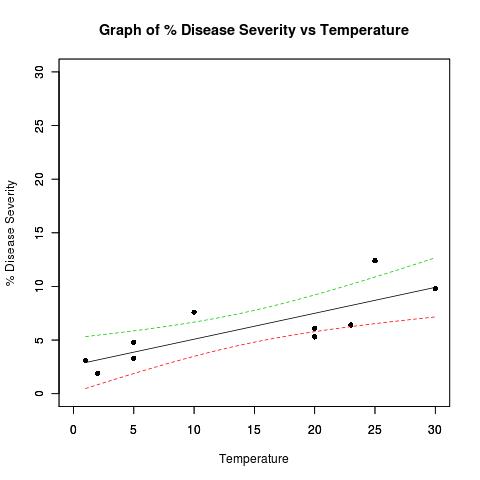
## Dataset from http://www.apsnet.org/education/advancedplantpath/topics/RModules/doc1/04_Linear_regression.html
## Disease severity as a function of temperature
# Response variable, disease severity
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
# Take a look at the data
plot(
diseasesev~temperature,
data=severity,
xlab="Temperature",
ylab="% Disease Severity",
pch=16,
pty="s",
xlim=c(0,30),
ylim=c(0,30)
)
title(main="Graph of % Disease Severity vs Temperature")
par(new=TRUE) # don't start a new plot
## Get datapoints predicted by best fit line and confidence bands
## at every 0.01 interval
xRange=data.frame(temperature=seq(min(temperature),max(temperature),0.01))
pred4plot <- predict(
lm(diseasesev~temperature),
xRange,
level=0.95,
interval="confidence"
)
## Plot lines derrived from best fit line and confidence band datapoints
matplot(
xRange,
pred4plot,
lty=c(1,2,2), #vector of line types and widths
type="l", #type of plot for each column of y
xlim=c(0,30),
ylim=c(0,30),
xlab="",
ylab=""
)
If I am interpreting confidence bands correctly, if a point does not lie within the confidence band it means that there is 95% chance that its not within the range of the mean predicted value for that specific x value (observed measurement) and nothing else (I cannot say anymore).
A confidence band is the lines on a scatter plot or fitted line plot that depict the upper and lower confidence bounds for all points on the range of data.
Well, I thought that this would be pretty easy with ggplot2, but now I realize that I have no idea how the confidence limits for stat_smooth/geom_smooth are calculated.
Consider the following:
library(ggplot2)
pred <- as.data.frame(predict(severity.lm,level=0.95,interval="confidence"))
dat <- data.frame(diseasesev,temperature,
in_interval = diseasesev <=pred$upr & diseasesev >=pred$lwr ,pred)
ggplot(dat,aes(y=diseasesev,x=temperature)) +
stat_smooth(method='lm') + geom_point(aes(colour=in_interval)) +
geom_line(aes(y=lwr),colour=I('red')) + geom_line(aes(y=upr),colour=I('red'))
This produces: alt text http://ifellows.ucsd.edu/pmwiki/uploads/Main/strangeplot.jpg
I don't understand why the confidence band calculated by stat_smooth is inconsistent with the band calculated directly from predict (i.e. the red lines). Can anyone shed some light on this?
Edit:
figured it out. ggplot2 uses 1.96 * standard error to draw the intervals for all smoothing methods.
pred <- as.data.frame(predict(severity.lm,se.fit=TRUE,
level=0.95,interval="confidence"))
dat <- data.frame(diseasesev,temperature,
in_interval = diseasesev <=pred$fit.upr & diseasesev >=pred$fit.lwr ,pred)
ggplot(dat,aes(y=diseasesev,x=temperature)) +
stat_smooth(method='lm') +
geom_point(aes(colour=in_interval)) +
geom_line(aes(y=fit.lwr),colour=I('red')) +
geom_line(aes(y=fit.upr),colour=I('red')) +
geom_line(aes(y=fit.fit-1.96*se.fit),colour=I('green')) +
geom_line(aes(y=fit.fit+1.96*se.fit),colour=I('green'))
The easiest way is probably to calculate a vector of TRUE/FALSE values that indicate if a data point is inside of the confidence interval or not. I'm going to reshuffle your example a little bit so that all of the calculations are completed before the plotting commands are executed- this provides a clean separation in the program logic that could be exploited if you were to package some of this into a function.
The first part is pretty much the same, except I replaced the additional call to lm() inside predict() with the severity.lm variable- there is no need to use additional computing resources to recalculate the linear model when we already have it stored:
## Dataset from
# apsnet.org/education/advancedplantpath/topics/
# RModules/doc1/04_Linear_regression.html
## Disease severity as a function of temperature
# Response variable, disease severity
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
## Get datapoints predicted by best fit line and confidence bands
## at every 0.01 interval
xRange=data.frame(temperature=seq(min(temperature),max(temperature),0.01))
pred4plot <- predict(
severity.lm,
xRange,
level=0.95,
interval="confidence"
)
Now, we'll calculate the confidence intervals for the origional data points and run a test to see if the points are inside the interval:
modelConfInt <- predict(
severity.lm,
level = 0.95,
interval = "confidence"
)
insideInterval <- modelConfInt[,'lwr'] < severity[['diseasesev']] &
severity[['diseasesev']] < modelConfInt[,'upr']
Then we'll do the plot- first a the high-level plotting function plot(), as you used it in your example, but we will only plot the points inside the interval. We will then follow up with the low-level function points() which will plot all the points outside the interval in a different color. Finally, matplot() will be used to fill in the confidence intervals as you used it. However instead of calling par(new=TRUE) I prefer to pass the argument add=TRUE to high-level functions to make them act like low level functions.
Using par(new=TRUE) is like playing a dirty trick a plotting function- which can have unforeseen consequences. The add argument is provided by many functions to cause them to add information to a plot rather than redraw it- I would recommend exploiting this argument whenever possible and fall back on par() manipulations as a last resort.
# Take a look at the data- those points inside the interval
plot(
diseasesev~temperature,
data=severity[ insideInterval,],
xlab="Temperature",
ylab="% Disease Severity",
pch=16,
pty="s",
xlim=c(0,30),
ylim=c(0,30)
)
title(main="Graph of % Disease Severity vs Temperature")
# Add points outside the interval, color differently
points(
diseasesev~temperature,
pch = 16,
col = 'red',
data = severity[ !insideInterval,]
)
# Add regression line and confidence intervals
matplot(
xRange,
pred4plot,
lty=c(1,2,2), #vector of line types and widths
type="l", #type of plot for each column of y
add = TRUE
)
I liked the idea and tried to make a function for that. Of course it's far from being perfect. Your comments are welcome
diseasesev<-c(1.9,3.1,3.3,4.8,5.3,6.1,6.4,7.6,9.8,12.4)
# Predictor variable, (Centigrade)
temperature<-c(2,1,5,5,20,20,23,10,30,25)
## For convenience, the data may be formatted into a dataframe
severity <- as.data.frame(cbind(diseasesev,temperature))
## Fit a linear model for the data and summarize the output from function lm()
severity.lm <- lm(diseasesev~temperature,data=severity)
# Function to plot the linear regression and overlay the confidence intervals
ci.lines<-function(model,conf= .95 ,interval = "confidence"){
x <- model[[12]][[2]]
y <- model[[12]][[1]]
xm<-mean(x)
n<-length(x)
ssx<- sum((x - mean(x))^2)
s.t<- qt(1-(1-conf)/2,(n-2))
xv<-seq(min(x),max(x),(max(x) - min(x))/100)
yv<- coef(model)[1]+coef(model)[2]*xv
se <- switch(interval,
confidence = summary(model)[[6]] * sqrt(1/n+(xv-xm)^2/ssx),
prediction = summary(model)[[6]] * sqrt(1+1/n+(xv-xm)^2/ssx)
)
# summary(model)[[6]] = 'sigma'
ci<-s.t*se
uyv<-yv+ci
lyv<-yv-ci
limits1 <- min(c(x,y))
limits2 <- max(c(x,y))
predictions <- predict(model, level = conf, interval = interval)
insideCI <- predictions[,'lwr'] < y & y < predictions[,'upr']
x_name <- rownames(attr(model[[11]],"factors"))[2]
y_name <- rownames(attr(model[[11]],"factors"))[1]
plot(x[insideCI],y[insideCI],
pch=16,pty="s",xlim=c(limits1,limits2),ylim=c(limits1,limits2),
xlab=x_name,
ylab=y_name,
main=paste("Graph of ", y_name, " vs ", x_name,sep=""))
abline(model)
points(x[!insideCI],y[!insideCI], pch = 16, col = 'red')
lines(xv,uyv,lty=2,col=3)
lines(xv,lyv,lty=2,col=3)
}
Use it like this:
ci.lines(severity.lm, conf= .95 , interval = "confidence")
ci.lines(severity.lm, conf= .85 , interval = "prediction")
If you love us? You can donate to us via Paypal or buy me a coffee so we can maintain and grow! Thank you!
Donate Us With